Addressing Variations and Uncertainties in Medical Image Data for Deep Learning
1. The Vessel Imbalance Problem in Retinal Vessel Segmentation

The vessel imbalance problem and the proposed three-stage deep learning framework.
Automatic retinal vessel segmentation is a fundamental step in the diagnosis of eye-related diseases, in which both thick vessels and thin vessels are important features for symptom detection. All existing deep learning models attempt to segment both types of vessels simultaneously by using a unified pixelwise loss which treats all vessel pixels with equal importance. Due to the highly imbalanced ratio between thick vessels and thin vessels (namely the majority of vessel pixels belong to thick vessels), the pixel-wise loss would be dominantly guided by thick vessels and relatively little influence comes from thin vessels, often leading to low segmentation accuracy for thin vessels. To address the imbalance problem, we explore to segment thick vessels and thin vessels separately by proposing a three-stage deep learning model. The vessel segmentation task is divided into three stages, namely thick vessel segmentation, thin vessel segmentation and vessel fusion. As better discriminative features could be learned for separate segmentation of thick vessels and thin vessels, this process minimizes the negative influence caused by their highly imbalanced ratio. The final vessel fusion stage refines the results by further identifying non-vessel pixels and improving the overall vessel thickness consistency. The experiments on public datasets DRIVE, STARE and CHASE DB1 clearly demonstrate that the proposed three-stage deep learning model outperforms the current state-of-theart vessel segmentation methods.
Publication:
Z. Yan, X. Yang, and K. -T. Cheng. A three-stage deep learning model for accurate retinal vessel segmentation. IEEE J. Biomed. Health Inform., 23(4):1427-1436, 2019.
2. The Inter-Observer Problem in Retinal Vessel Segmentation
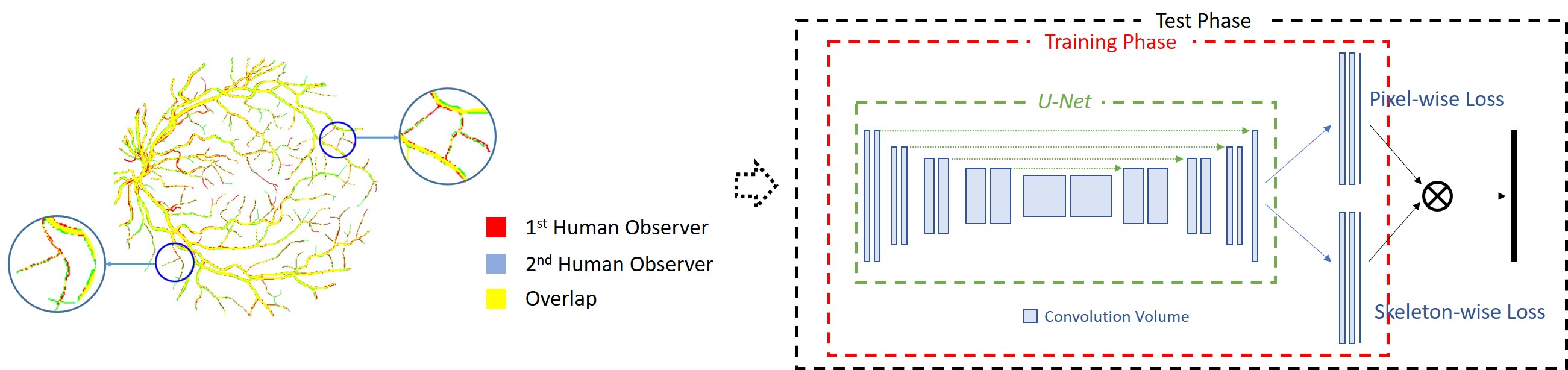
The inter-observer problem and the corresponding proposed joint-loss deep learning framework.
Limited by the resolution of fundus image, manually annotated vessels in different annotations are often not perfectly matched. For the basic vessel structure (or thick vessels), locations annotated by di erent observers are quite similar, but vessel thickness could be slightly different. Though the thickness variation is limited, their influence can be disproportional due to the highly imbalanced ratio between the basic vessel structure and tiny vessels. In different manual annotations made to the same fundus image, vessel thickness of tiny vessels is usually similar (not necessarily the same) but their locations can be quite different. In other words, for tiny vessels, location variation in different annotations can be quite serious. Based on the analysis of the entire dataset, we found that the inter-observer problem is quite common and it is not unusual at all that non-vessel pixels in one manual annotation can be vessel pixels in another manual annotation. Therefore, the pixel-to-pixel matching strategy should not be directly used for constructing loss functions, and a good loss function must tolerate such variations caused by the inter-observer problem. Therefore, we propose a skeleton-level shape similairty measure and design a loss function for the segmentation of thin vessels. The proposed joint-loss deep learning framework is able to bring consistent performance improvement for both deep and shallow network architectures, achieving the best overall performance on public datasets.
Publication:
Z. Yan, X. Yang, and K. -T. Cheng. A skeletal similarity metric for quality evaluation of retinal vessel segmentation. IEEE Trans. Med. Imag., 37(4):1045-1057, 2018.
Z. Yan, X. Yang, and K. -T. Cheng. Joint segment-level and pixel-wise losses for deep learning based retinal vessel segmentation. IEEE Trans. Biomed. Eng., 65(9):1912-1923, 2018.
3. The Boundary Uncertainty Problem in Gland Instance Segmentation
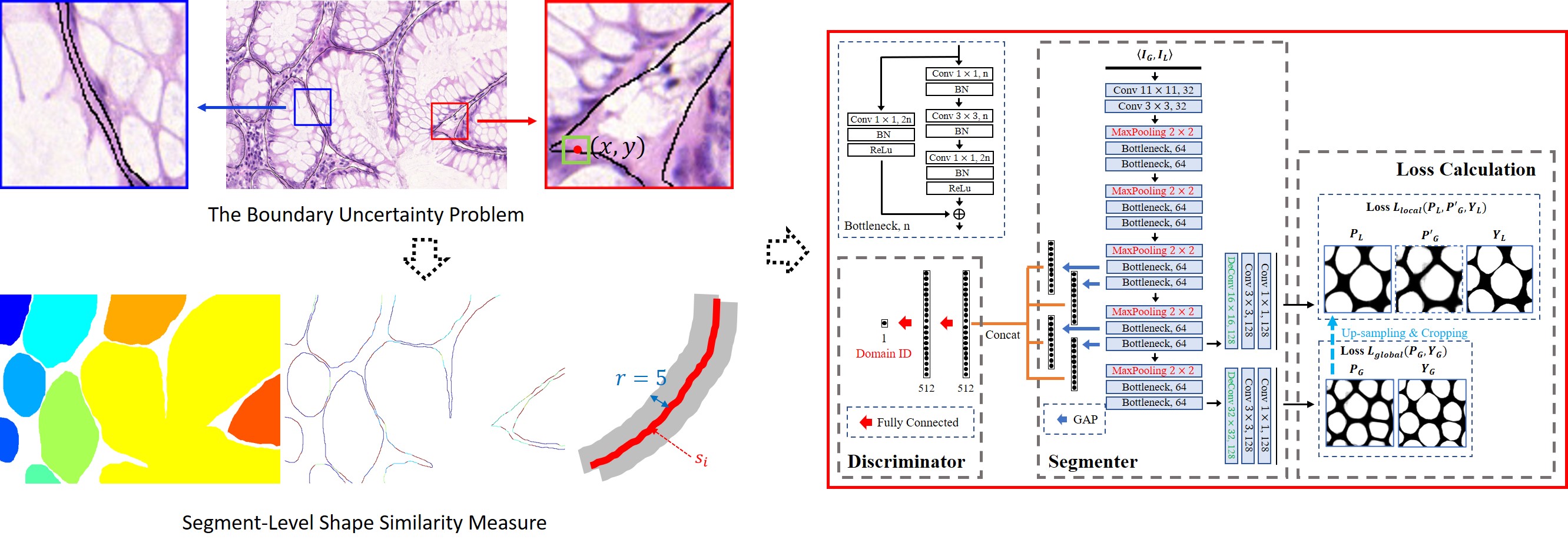
The boundary uncertainty problem and the corresponding proposed shape-aware adversarial learning framework.
Segmenting gland instances in histology images is highly challenging as it requires not only detecting glands from a complex background but also separating each individual gland instance with accurate boundary detection. However, due to the boundary uncertainty problem in manual annotations, pixel-to-pixel matching based loss functions are too restrictive for simultaneous gland detection and boundary detection. State-of-the-art approaches adopted multi-model schemes, resulting in unnecessarily high model complexity and difficulties in the training process. We propose to use one single deep learning model for accurate gland instance segmentation. To address the boundary uncertainty problem, instead of pixel-to-pixel matching, we propose a segment-level shape similarity measure to calculate the curve similarity between each annotated boundary segment and the corresponding detected boundary segment within a fixed searching range. As the segment-level measure allows location variations within a fixed range for shape similarity calculation, it has better tolerance to boundary uncertainty and is more effective for boundary detection. Furthermore, by adjusting the radius of the searching range, the segment-level shape similarity measure is able to deal with different levels of boundary uncertainty. Therefore, in our framework, images of different scales are down-sampled and integrated to provide both global and local contextual information for training, which is helpful in segmenting gland instances of different sizes. To reduce the variations of multi-scale training images, by referring to adversarial domain adaptation, we propose a pseudo domain adaptation framework for feature alignment. By constructing loss functions based on the segment-level shape similarity measure, combining with the adversarial loss function, the proposed shape-aware adversarial learning framework enables one single deep learning model for gland instance segmentation. Experimental results on the 2015 MICCAI Gland Challenge dataset demonstrate that the proposed framework achieves state-of-the-art performance with one single deep learning model. As the boundary uncertainty problem widely exists in medical image segmentation, it is broadly applicable to other applications.
Publication:
Z. Yan, X. Yang, and K. -T. Cheng. A deep model with shape-preserving loss for gland instance segmentation. in The International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), pages 138-146, 2018.
Z. Yan, X. Yang, and K. -T. Cheng. Enabling a single deep learning model for accurate gland instance segmentation: A shape-aware adversarial learning framework. IEEE Trans. Med. Imag., 39(6):2176-2189, 2020.
4. The Cross-Client Variation Problem in Federated Learning with Multi-Source Decentralized Medical Image Data
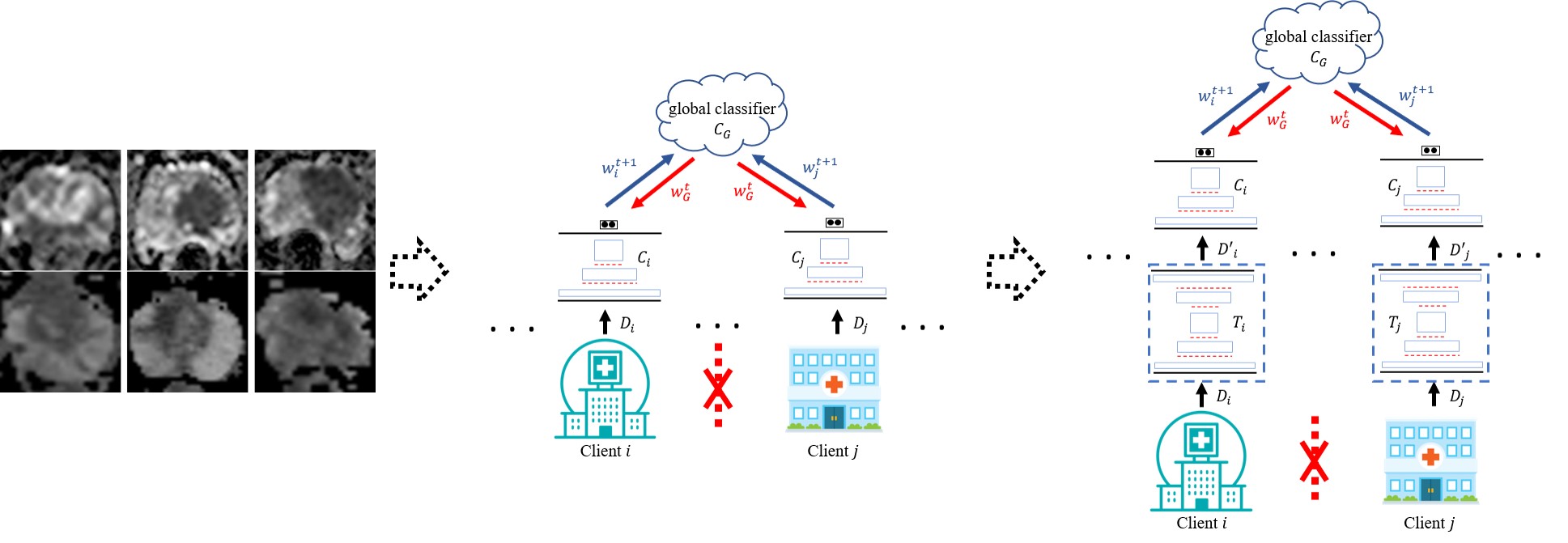
The cross-client variation problem and the corresponding proposed variation-aware federated learning framework.
Deep learning approaches for medical image analysis have been limited by the lack of large datasets. Privacy concerns, especially for medical image data, make it almost infeasible to construct a large dataset by fusing multiple small ones from different sources for training deep learning models. Therefore, federated learning (FL), which has the potential for training deep learning models from multi-source decentralized data with privacy preservation, has drawn great attention recently. However, as the amount of annotated medical image data available at each client (i.e. a hospital or a medical center) usually is quite limited, parameters updated at each client in FL can be highly sensitive to its local data. As a result, even small variations among medical image data across clients would lead to significant performance degradation. We, for the first time, take the cross-client variation problem in FL into consideration and propose a variation-aware federated learning (VAFL) framework. In contrast to typical horizontal FL where the parameters of each client are directly updated based on the client’s raw training images, VAFL updates the parameters based on transformed images through image-to-image translation. By transforming the images of all clients onto a common image space, the variations among clients can be effectively minimized. Specifically, we first select one client whose training images are most suitable to be used for defining the target image space. Instead of sharing raw images among clients which is prohibited due to privacy concerns, a small set of images are synthesized based on the chosen client’s images and further shared with all other clients which are used by each client for training an image-to-image translator for each individual client. The synthesized images capture the key features to represent the target space with high fidelity while they are sufficiently distinct from any raw image from the selected client. Therefore, the cross-client variation problem is addressed with privacy preservation. In our case study, we apply the framework for automated classification of clinically significant prostate cancer and evaluate it using multi-source decentralized apparent diffusion coefficient (ADC) image data. Experimental results demonstrate that the proposed VAFL framework stably outperforms the current horizontal FL framework. In addition, we discuss the conditions, and experimentally validated them, that VAFL is applicable for training a global model among multiple clients instead of directly training deep learning models locally on each client. Checking the satisfiability of such conditions can be used as guidance in determining if VAFL or FL should be employed for multi-source decentralized medical image data.
Publication:
Z. Yan, J. Wicaksana, Z. Wang, X. Yang, and K. -T. Cheng. Variation-aware federated learning with multi-source decentralized medical image data. IEEE J. Biomed. Health Inform., 2020.